Biography
叶盛博士,现任安徽大学人工智能学院教授,安徽省优青,安徽省无人系统与智能技术工程研究中心副主任,安徽省通用人工智能领域战略咨询专家,VIEW期刊青年编委。2021年获中国科学技术大学化学物理系博士学位(导师江俊教授),中科院百篇优博获得者,安徽省省域拔尖人才,合肥市市级领军人才。先后获得过中国科学院优秀博士学位论文、中科院院长奖“特别奖”、中国化学会京博科技奖优秀博士论文奖、中国科学技术大学优秀博士毕业论文,中国科学技术大学优秀毕业生、博士研究生国家奖学金等荣誉奖励。作为项目负责人先后主持安徽省自然科学基金优青项目,国家自然科学基金面上、青年项目,安徽省高校与合肥综合性国家科学中心协同创新项目。
Dr. Sheng Ye was born in Anhui, China, in 1994, received his B.S. degree in Materials Science and Engineering from Anhui University of Science and Technology in 2016 and earned his Ph.D. in Theoretical Chemistry under the supervision of Prof. Jun Jiang at the University of Science and Technology of China in 2021. Since then, he has served as a professor at the School of Artificial Intelligence at Anhui University, focusing on Theoretical Chemistry, integrating Machine Learning into his research. In 2022, Sheng was recognized as an Excellent Young Scholar in Anhui Province. He also serves as a strategic consultant in the field of general artificial intelligence for Anhui Province and is a youth editorial board member of the journal VIEW.
Honor
Dr. Ye has received numerous accolades, including the prestigious “100 Excellent Doctoral Dissertations of the Chinese Academy of Sciences (CAS)” award. He has been recognized as a Youth Talent of Anhui Province, an Outstanding Talent of Anhui Province, and a Leading Talent of Hefei City. In addition to his role as a professor, Dr. Ye serves as Deputy Director of the Anhui Provincial Engineering Research Center for Unmanned Systems and Intelligent Technology and as a strategic consultant for Anhui Province in the area of General Artificial Intelligence Throughout his career, Dr. Ye has been honored with multiple distinguished awards, such as the CAS Excellent Doctoral Dissertation Award, the CAS President’s Award “Special Award,” the Excellent Doctoral Dissertation Award from the Chinese Chemical Society’s Jingbo Technology Award, the Outstanding Doctoral Dissertation Award from the University of Science and Technology of China (USTC), the Outstanding Graduate Award from USTC, and the National Scholarship for Doctoral Students.
Research Profile
叶盛博士的研究主要聚焦人工智能与量子化学的前沿交叉研究,立足物理化学基本规律,结合机器学习与多尺度理论方法,面向功能分子、复合材料以及生物体系中的蛋白与药物,开展构效关系解析与逆向从头设计研究,揭示复杂体系的微观机制与协同原理。系统性地发展了结合量子化学与人工智能的蛋白质光谱模拟方法,以及基于物理“谱-构-效”关系的多功能蛋白质从头设计方法。近五年以第一作者或通讯作者在国际知名期刊如PNAS, Natl Sci Rev, JACS, JACS Au, JPCL, and Energy & Environmental Science 等发表论文二十余篇,并且多项研究工作被国际顶级学术期刊Science (Science, 2020, 370, 1178; Science, 2020, 368, 727) 专文推荐点评,研究成果受到了国际上的广泛认可。所开发的蛋白质光谱人工智能模拟软件目前已在中国科学技术大学、南京大学、南欣医药研究院公司、美国加州大学尔湾分校、英国诺丁汉大学、英国杜伦大学、意大利INFN国家实验室等十几家单位应用。
Dr. Sheng Ye's research primarily focuses on the intersection of artificial intelligence and quantum chemistry, aiming to address practical challenges across various applications in physics and chemistry, such as protein inverse design, molecular inverse design, molecular spectroscopy simulation, and energy catalytic materials. He has systematically developed protein spectroscopy simulation methods that integrate quantum chemistry with AI and proposed a high-throughput, de novo protein design approach based on the physics-driven spectrum-structure-function relationship. Over the past five years, he has published 17 papers in prestigious journals such as PNAS, Natl Sci Rev, JACS, JACS Au, JPCL, and Energy & Environmental Science. Additionally, his research has been featured twice as an "Editor's Choice" in Science (Science, 2020, 370, 1178; Science, 2020, 368, 727).The AI-driven protein spectroscopy simulation software developed by Dr. Ye has currently been applied at over a dozen institutions, including the University of Science and Technology of China, Nanjing University, Naxin Pharmaceutical Research Institute, University of California, Irvine, University of Nottingham, Durham University, and Italy's INFN National Laboratory.
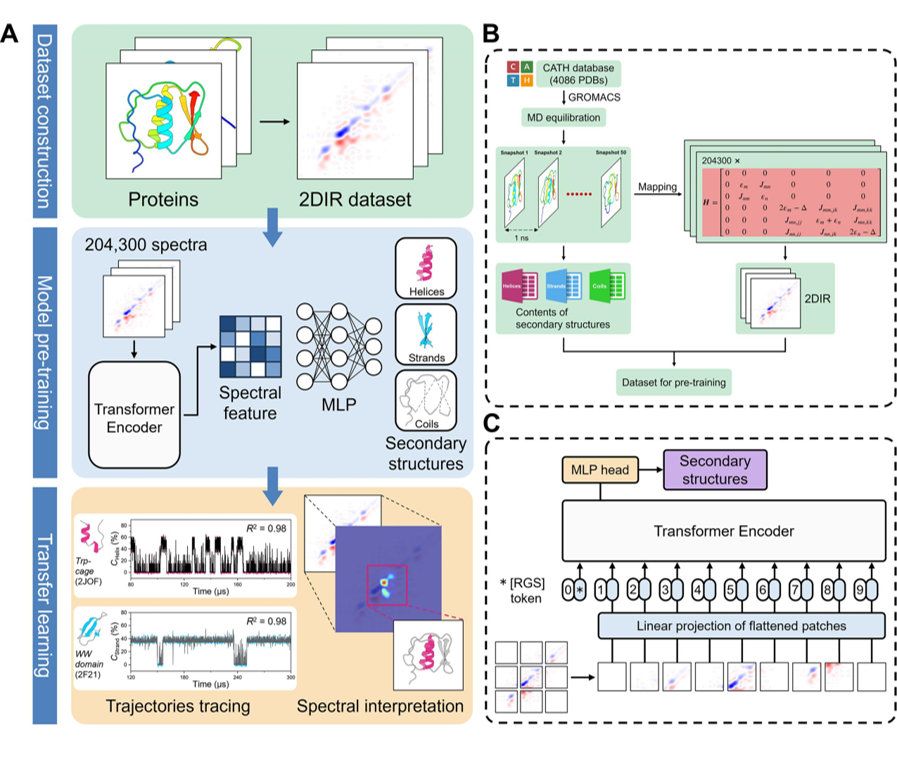
Unraveling dynamic protein structures from two-dimensional infrared (2DIR) spectral signals presents a formidable task. By utilizing machine learning, we have correlated the intricate 2DIR spectroscopic features with the protein dynamic conformations, thereby tracing changes of secondary structure content along protein folding trajectories. This pretrained model facilitates universal transfer learning across diverse protein folding trajectories, offering valuable insights into the dynamic behavior of proteins and their biological function.
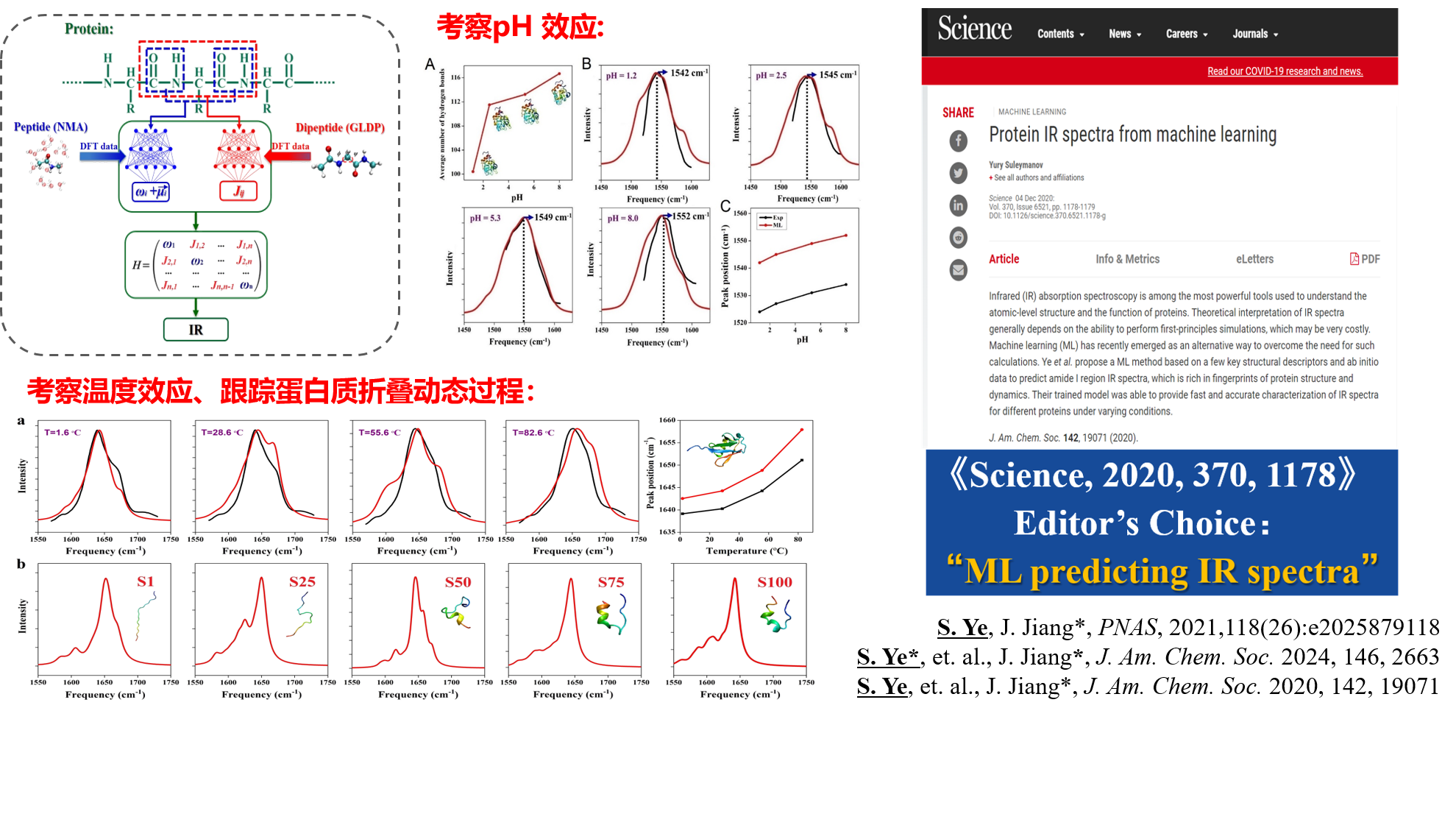
By combining machine learning techniques with quantum chemical theoretical calculations, we have developed a divide-and-conquer artificial intelligence simulation method for protein spectroscopy, along with the corresponding simulation software, MLPS (http://dcaiku.com:12880/platform/first). This tool enables efficient and accurate simulation of the amide I and II bands in protein infrared spectra, precisely distinguishing protein secondary structures. It can also be used to investigate the effects of temperature and solution pH on protein structures and to track the protein folding process in real-time.
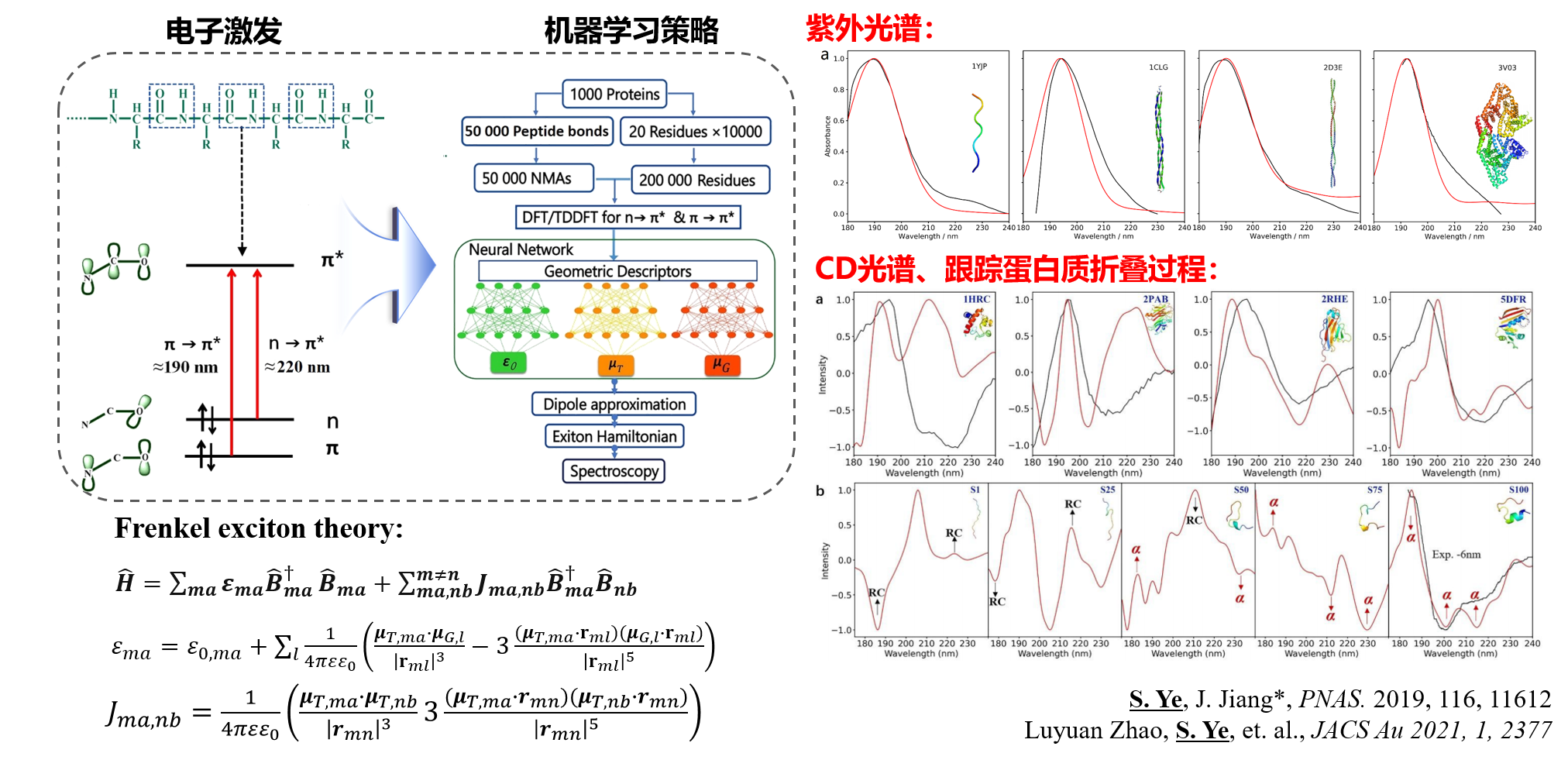
We have integrated the Frenkel exciton model to develop a divide-and-conquer machine learning prediction strategy for protein CD and UV spectra. This method improves computational speed by at least three orders of magnitude compared to traditional quantum chemical calculations. Furthermore, the model demonstrates excellent transferability, making it applicable to simulating photoelectric responses under various external conditions (such as temperature, pressure, and electric fields). It can also be employed to predict the circular dichroism (CD) spectra of dynamic conformations during the Trp-cage protein folding process, enabling real-time monitoring of conformational changes in protein folding dynamics.
New Progress
🎉 Our paper, “A Machine Learning Protocol for Predicting Structural Distributions of Amyloid-Forming Proteins from 2D IR Spectra” (S. Ye*, et al) has been accepted for publication in Proc. Natl. Acad. Sci. U.S.A. 2025!
🎉 Our review paper, “人工智能驱动的蛋白质结构预测进展与应用前景” (S. Ye*, et al) has been accepted for publication in JOURNAL OF BIOLOGY 2025!
🎉 Our paper, “AI Protocol for Retrieving Protein Dynamic Structures from Two-Dimensional Infrared Spectra” (S. Ye*, et al) has been accepted for publication in Proc. Natl. Acad. Sci. U.S.A. 2025!
🎉 Sheng was selected as a youth editorial board member of the journal VIEW (JCR 一区, IF:9.7)!
🎉 Our paper, “Monitoring C-C coupling in catalytic reactions via machine-learned infrared spectroscopy” (S. Ye*, et al) has been accepted for publication in National Science Review!
🎉 Our paper, “Unraveling dynamic protein structures by two-dimensional infrared spectra with a pretrained machine learning model” (S. Ye*, et al) has been accepted for publication in Proc. Natl. Acad. Sci. U.S.A. 2024, 121 (27), e2409257121.
🎉 Our paper, “Artificial Intelligence-based Amide-II Infrared Spectroscopy Simulation for Monitoring Protein Hydrogen Bonding Dynamics” (S. Ye*, et al) has been accepted for publication in J. Am. Chem. Soc. 2024, 146 (4), 2663-2672.
🎉 Our paper, “Machine Learning Prediction of Molecular Binding Profiles on Metal-Porphyrin via Spectroscopic Descriptors” (S. Ye*, et al) has been accepted for publication in J. Phys. Chem. Lett. 2024, 15 (7), 1956-1961.
🎉 Our paper, “Catalytic Structure Design by AI Generating with Spectroscopic Descriptors” (S. Ye (co-first author), et al) has been accepted for publication in J. Am. Chem. Soc. 2023, 145 (49), 26817-26823.
🎉 Our paper, “High-throughput oxygen chemical potential engineering of perovskite oxides for chemical looping applications” has been accepted for publication in Energy Environ. Sci. 2022, 15 (4), 1512-1528.
🎉 Our paper, “AI-based spectroscopic monitoring of real-time interactions between SARS-CoV-2 and human ACE2” (S. Ye, …, J. Jiang*) has been accepted for publication in Proc. Natl. Acad. Sci. U.S.A. 2021, 118 (26), e2025879118.
🎉 Our paper, “A Machine Learning Protocol for Predicting Protein Infrared Spectra” (S. Ye, …, J. Jiang*) has been accepted for publication in J. Am. Chem. Soc. 142 (2020) 19071-19077. 审稿人评价本工作 “It represents a significant advance in the field and offers a new framework to understand the protein structure and folding aided by artificial intelligence techniques”, 并赞扬我们 “present an innovative approach that will be of value to a broad community of computational spectroscopists.” 2020年12月,Science杂志编辑撰稿专文推荐我们发展的红外光谱机器学习预测方法(Science 2020, 370, 1178),指出“ Their trained model was able to provide fast and accurate characterization of IR spectra for different proteins under varying conditions.”
🎉 Our paper, “Electric Dipole Descriptor for Machine Learning Prediction of Catalyst Surface-Molecular Adsorbate Interactions” (X.J. Wang, S. Ye (co-first author), et al) has been accepted for publication in J. Am. Chem. Soc. 142 (2020) 7737-7743. 审稿人评价本工作 “The findings are novel and interesting, the simulations and neural network protocol are state-of-the-art”, 并指出 “It could be very useful for the design and evaluation of new inhomogeneous catalysts.” 该工作于2020-5-19日被国家自然科学基金委网站作为重要资助成果报道。2020年5月,Science杂志编辑撰稿专文推荐我们首创的电偶极矩相关描述符(Science 2020, 368, 727),指出 “The transferability of these models between different heterogeneous systems suggests that the electric dipole moment is a promising new type of catalytic descriptor.”
🎉 Our paper, “A neural network protocol for electronic excitations of N-methylacetamide” (S. Ye, …, J. Jiang*) has been accepted for publication in Proc. Natl. Acad. Sci. U.S.A. 2019, 116 (24), 11612-11617.
Publications
(19) Ye, S, et al, A Machine Learning Protocol for Predicting Structural Distributions of Amyloid-Forming Proteins from 2D IR Spectra Proc. Natl. Acad. Sci. U.S.A., 2025, in press.
(18) Ye, S, L. Zhu, Z. Zhao, F. Wu, Z. Li, B. Wang, K. Zhong, C. Sun, S. Mukamel & J. Jiang. AI Protocol for Retrieving Protein Dynamic Structures from Two-Dimensional Infrared Spectra. Proc. Natl. Acad. Sci. U.S.A., 2025, 122 (7) e2424078122.
(17) Li Yang, Zhicheng Zhao, Tongtong Yang, Donglai Zhou, Xiaoyu Yue, Xiyu Li, Yan Huang, X. Wang, Ruyun Zheng, Thomas Heine, Changyin Sun, Jun Jiang,* and Ye, S.* (Corresponding Author). Monitoring C-C coupling in catalytic reactions via machine-learned infrared spectroscopy. Natl Sci Rev, 2025, 12: nwae389.
(16) Ye, S * (First author and corresponding author).; Zhong, K.; Huang, Y.; Zhang, G.; Sun, C.; Jiang, J., Artificial Intelligence-based Amide-II Infrared Spectroscopy Simulation for Monitoring Protein Hydrogen Bonding Dynamics. J. Am. Chem. Soc. 2024, 146 (4), 2663-2672.
(15) Wu, F.; Huang, Y.; Yang, G.; Ye, S.* (Corresponding Author); Mukamel, S.; Jiang, J., Unraveling dynamic protein structures by two-dimensional infrared spectra with a pretrained machine learning model. Proc. Natl. Acad. Sci. U.S.A. 2024, 121 (27), e2409257121.
(14) Ye, S.; Zhong, K.; Zhang, J.; Hu, W.; Hirst, J. D.; Zhang, G.; Mukamel, S.; Jiang, J., A machine learning protocol for predicting protein infrared spectra. J. Am. Chem. Soc. 2020, 142 (45), 19071-19077.
(13) Ye, K.; Wang, S.; Huang, Y.; Hu, M.; Zhou, D.; Luo, Y.; Ye, S.* (Corresponding Author); Zhang, G.; Jiang, J., Machine Learning Prediction of Molecular Binding Profiles on Metal-Porphyrin via Spectroscopic Descriptors. J. Phys. Chem. Lett. 2024, 15 (7), 1956-1961.
(12) Ye, S.; Hu, W.; Li, X.; Zhang, J.; Zhong, K.; Zhang, G.; Luo, Y.; Mukamel, S.; Jiang, J., A neural network protocol for electronic excitations of N-methylacetamide. Proc. Natl. Acad. Sci. U.S.A. 2019, 116 (24), 11612-11617.
(11) Ye, S.; Zhang, G.; Jiang, J., AI-based spectroscopic monitoring of real-time interactions between SARS-CoV-2 and human ACE2. Proc. Natl. Acad. Sci. U.S.A. 2021, 118 (26), e2025879118.
(10) Yang, T.; Zhou, D.; Ye, S. (co-first author); Li, X.; Li, H.; Feng, Y.; Jiang, Z.; Yang, L.; Ye, K.; Shen, Y., Catalytic Structure Design by AI Generating with Spectroscopic Descriptors. J. Am. Chem. Soc. 2023, 145 (49), 26817-26823.
(9) Wang, X.; Ye, S. (co-first author); Hu, W.; Sharman, E.; Liu, R.; Liu, Y.; Luo, Y.; Jiang, J., Electric dipole descriptor for machine learning prediction of catalyst surface–molecular adsorbate interactions. J. Am. Chem. Soc. 2020, 142 (17), 7737-7743.
(8) Hu, W.; Ye, S (co-first author).; Zhang, Y.; Li, T.; Zhang, G.; Luo, Y.; Mukamel, S.; Jiang, J., Machine learning protocol for surface-enhanced Raman spectroscopy. J. Phys. Chem. Lett. 2019, 10 (20), 6026-6031.
(7) Zhang, Y.; Ye, S (co-first author).; Zhang, J.; Hu, C.; Jiang, J.; Jiang, B., Efficient and Accurate Spectroscopic Simulations with Symmetry-Preserving Neural Network Models for Tensorial Properties. J. Phys. Chem. B 2020, 124 (33), 7284-7290.
(6) Zhang, J.; Ye, S (co-first author).; Zhong, K.; Zhang, Y.; Chong, Y.; Zhao, L.; Zhou, H.; Guo, S.; Zhang, G.; Jiang, B., A machine-learning protocol for ultraviolet protein-backbone absorption spectroscopy under environmental fluctuations. J. Phys. Chem. B 2021, 125 (23), 6171-6178.
(5) Zhao, L.; Zhang, J.; Zhang, Y.; Ye, S.; Zhang, G.; Chen, X.; Jiang, B.; Jiang, J., Accurate machine learning prediction of protein circular dichroism spectra with embedded density descriptors. JACS Au 2021, 1 (12), 2377-2384.
(4) Wang, X.; Gao, Y.; Krzystowczyk, E.; Iftikhar, S.; Dou, J.; Cai, R.; Wang, H.; Ruan, C.; Ye, S.; Li, F., High-throughput oxygen chemical potential engineering of perovskite oxides for chemical looping applications. Energy Environ. Sci. 2022, 15 (4), 1512-1528.
(3) Yu, H.; Wang, Y.; Wang, X.; Zhang, J.; Ye, S.; Huang, Y.; Luo, Y.; Sharman, E.; Chen, S.; Jiang, J., Using machine learning to predict the dissociation energy of organic carbonyls. J. Phys. Chem. A 2020, 124 (19), 3844-3850.
(2) Wang, X.; Jiang, S.; Hu, W.; Ye, S.; Wang, T.; Wu, F.; Yang, L.; Li, X.; Zhang, G.; Chen, X., Quantitatively determining surface–adsorbate properties from vibrational spectroscopy with interpretable machine learning. J. Am. Chem. Soc. 2022, 144 (35), 16069-16076.
(1) Feng, C.; Sharman, E.; Ye, S.; Luo, Y.; Jiang, J., A neural network protocol for predicting molecular bond energy. Sci. China Chem. 2019, 62, 1698-1703.
Academic Conference
- Sheng joined in the Shuangqing Forum of NSFC and gave a short talk, at Beijing.
- Sheng delivered an invited talk in the 15th National Conference on Theoretical and Computational Chemistry of the Chinese Chemical Society, at Changchun.
- Sheng delivered an invited talk in the The 34th Annual Academic Conference of the Chinese Chemical Society: Electronic structure theory and its applications, at Guangzhou.
- Sheng delivered an invited talk in the The 33th Annual Academic Conference of the Chinese Chemical Society: Frontiers in Physical Chemistry, at Qingdao.
